|
chr2_-_21084077
|
3.071
|
|
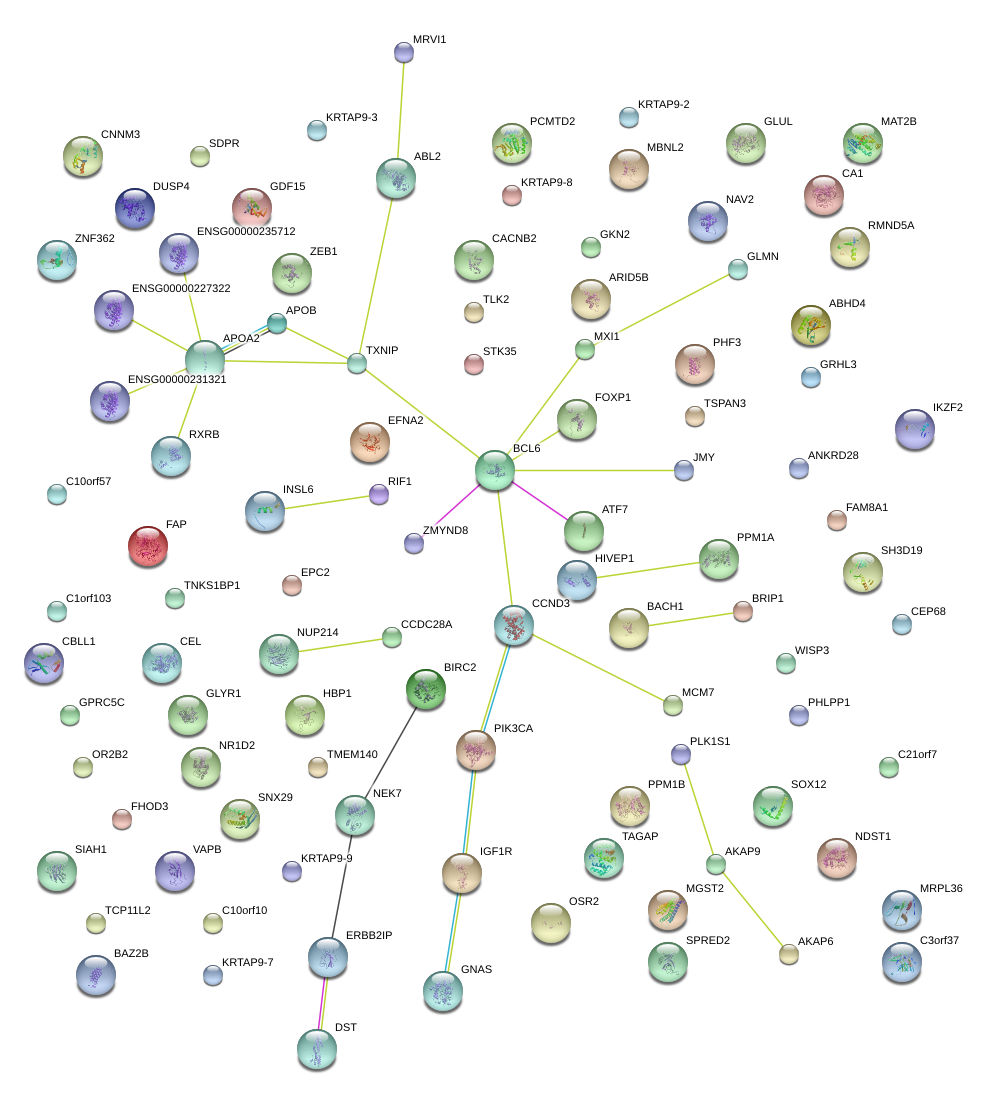
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr5_+_78568094
|
2.980
|
|
|
|
|
chr5_+_78567604
|
2.905
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr10_+_63478974
|
2.847
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr1_+_144149794
|
2.714
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr20_-_45410033
|
2.655
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr7_+_106596641
|
2.559
|
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr2_-_21084911
|
2.333
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr7_+_134483305
|
2.305
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr19_+_18357966
|
2.248
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr19_+_1158129
|
2.099
|
|
EFNA2
|
ephrin-A2
|
|
chr13_+_96672705
|
1.903
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr1_+_196392513
|
1.829
|
NM_133494
|
NEK7
|
NIMA (never in mitosis gene a)-related kinase 7
|
|
chr14_+_31868229
|
1.803
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr20_+_254205
|
1.792
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr5_+_90711887
|
1.739
|
|
LOC100129716
|
hypothetical LOC100129716
|
|
chr2_+_96845704
|
1.718
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr6_+_64403631
|
1.705
|
|
PHF3
|
PHD finger protein 3
|
|
chr6_+_64403685
|
1.691
|
|
PHF3
|
PHD finger protein 3
|
|
chr1_-_177378801
|
1.676
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr6_+_64403936
|
1.665
|
|
PHF3
|
PHD finger protein 3
|
|
chr4_+_140806536
|
1.643
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr6_+_64403668
|
1.577
|
|
PHF3
|
PHD finger protein 3
|
|
chr12_+_105220638
|
1.555
|
NM_152772
|
TCP11L2
|
t-complex 11 (mouse)-like 2
|
|
chr3_+_180348818
|
1.544
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr3_-_15814678
|
1.480
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_+_64414389
|
1.480
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr20_+_2030527
|
1.458
|
NM_080836
|
STK35
|
serine/threonine kinase 35
|
|
chr10_+_111959978
|
1.439
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr20_+_2031418
|
1.369
|
|
STK35
|
serine/threonine kinase 35
|
|
chr4_+_140806371
|
1.364
|
NM_002413
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr16_-_46957284
|
1.285
|
NM_001006610
|
SIAH1
|
seven in absentia homolog 1 (Drosophila)
|
|
chr20_+_62357490
|
1.282
|
NM_001104925
NM_018257
|
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2
|
|
chr2_+_86800940
|
1.271
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr17_+_69939130
|
1.259
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr6_+_17708483
|
1.251
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr3_+_23962518
|
1.234
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr16_-_46957254
|
1.233
|
|
SIAH1
|
seven in absentia homolog 1 (Drosophila)
|
|
chr3_-_71196696
|
1.231
|
|
FOXP1
|
forkhead box P1
|
|
chr7_+_91408086
|
1.231
|
NM_005751
NM_147185
|
AKAP9
|
A kinase (PRKA) anchor protein (yotiao) 9
|
|
chr2_+_86800898
|
1.229
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr20_+_21054623
|
1.227
|
NM_001163022
NM_001163023
NM_018474
|
PLK1S1
|
polo-like kinase 1 substrate 1
|
|
chr8_-_29263714
|
1.199
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr15_+_22781292
|
1.196
|
|
PAR5
|
Prader-Willi/Angelman syndrome-5
|
|
chr2_+_44249487
|
1.180
|
NM_001033556
NM_001033557
NM_002706
NM_177968
NM_177969
|
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B
|
|
chr2_+_86801081
|
1.174
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr3_-_188935363
|
1.174
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_-_69033515
|
1.165
|
NM_182536
|
GKN2
|
gastrokine 2
|
|
chr17_+_57909979
|
1.148
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr2_+_44249565
|
1.133
|
|
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B
|
|
chr5_+_149845503
|
1.125
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr1_+_33494745
|
1.108
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr3_-_71376919
|
1.105
|
|
FOXP1
|
forkhead box P1
|
|
chr3_+_23961739
|
1.098
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr20_-_42561725
|
1.072
|
|
|
|
|
chr6_+_112481970
|
1.044
|
NM_003880
NM_198239
|
WISP3
|
WNT1 inducible signaling pathway protein 3
|
|
chr6_+_12120709
|
1.043
|
NM_002114
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1
|
|
chr10_+_31650069
|
1.025
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_82163568
|
1.022
|
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr11_-_10630408
|
1.016
|
NM_001098579
|
MRVI1
|
murine retrovirus integration site 1 homolog
|
|
chr14_+_22136968
|
1.014
|
NM_022060
|
ABHD4
|
abhydrolase domain containing 4
|
|
chr18_+_58533597
|
1.003
|
NM_194449
|
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr6_-_159386007
|
0.995
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr15_-_75135496
|
0.979
|
|
TSPAN3
|
tetraspanin 3
|
|
chr10_+_63331018
|
0.978
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr10_-_44794146
|
0.968
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr6_+_139136349
|
0.962
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr11_+_20005704
|
0.951
|
|
NAV2
|
neuron navigator 2
|
|
chr8_+_100025806
|
0.948
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr6_+_134800533
|
0.939
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr6_-_56824371
|
0.931
|
|
DST
|
dystonin
|
|
chr11_+_101723125
|
0.927
|
|
BIRC2
|
baculoviral IAP repeat containing 2
|
|
chr2_-_65447370
|
0.922
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr6_-_27988152
|
0.917
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr1_-_159459814
|
0.888
|
|
APOA2
|
apolipoprotein A-II
|
|
chr2_-_160181206
|
0.886
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr2_+_65136989
|
0.877
|
NM_015147
|
CEP68
|
centrosomal protein 68kDa
|
|
chr9_+_132990760
|
0.874
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr12_-_52281050
|
0.873
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr7_+_107171514
|
0.872
|
NM_024814
|
CBLL1
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1
|
|
chr2_+_151974630
|
0.864
|
NM_001177663
NM_018151
NM_001177664
|
RIF1
|
RAP1 interacting factor homolog (yeast)
|
|
chr20_+_56397568
|
0.862
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr1_+_24518398
|
0.854
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr15_+_97009194
|
0.852
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr2_+_149119029
|
0.842
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr2_-_162808123
|
0.836
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr2_-_65447267
|
0.833
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr2_-_213724577
|
0.829
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr5_+_27508139
|
0.825
|
|
LOC643401
|
hypothetical protein LOC643401
|
|
chr6_-_33276131
|
0.821
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr14_+_59785723
|
0.813
|
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr3_+_64645819
|
0.812
|
|
LOC100507098
|
hypothetical LOC100507098
|
|
chr8_-_86441115
|
0.811
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr1_+_24518501
|
0.806
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr21_+_29593607
|
0.796
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr6_-_42124587
|
0.795
|
NM_001136017
NM_001136126
|
CCND3
|
cyclin D3
|
|
chr5_+_65258316
|
0.795
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr5_+_162865128
|
0.795
|
NM_013283
|
MAT2B
|
methionine adenosyltransferase II, beta
|
|
chr2_-_138975682
|
0.783
|
|
|
|
|
chr10_+_18729518
|
0.771
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr11_-_56846246
|
0.763
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr17_+_36642240
|
0.760
|
NM_031962
|
KRTAP9-9
KRTAP9-3
|
keratin associated protein 9-9
keratin associated protein 9-3
|
|
chr16_-_4836198
|
0.756
|
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr1_-_180624494
|
0.755
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr14_+_59785739
|
0.753
|
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr5_+_72287625
|
0.752
|
|
|
|
|
chr20_+_56904042
|
0.744
|
|
GNAS
|
GNAS complex locus
|
|
chr4_-_152368492
|
0.742
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr6_-_33276347
|
0.742
|
NM_021976
|
RXRB
|
retinoid X receptor, beta
|
|
chr7_-_99536214
|
0.728
|
NM_182776
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr2_-_192419896
|
0.728
|
|
SDPR
|
serum deprivation response
|
|
chr21_+_29439768
|
0.726
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr2_+_196230096
|
0.717
|
NM_020342
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr13_+_75232797
|
0.716
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr3_+_23904086
|
0.710
|
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast)
|
|
chr1_-_180624380
|
0.702
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr7_-_155129817
|
0.701
|
|
|
|
|
chr6_-_33276141
|
0.700
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr16_-_3362283
|
0.691
|
NM_001190476
|
MTRNR2L4
|
MT-RNR2-like 4
|
|
chr6_+_155579788
|
0.691
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr14_+_23937766
|
0.690
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr5_+_72287630
|
0.688
|
|
FCHO2
|
FCH domain only 2
|
|
chr5_+_137701852
|
0.686
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr5_+_137701855
|
0.679
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr1_-_150564302
|
0.679
|
NM_002016
|
FLG
|
filaggrin
|
|
chr2_+_172658453
|
0.676
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr5_+_72287509
|
0.675
|
NM_001146032
NM_138782
|
FCHO2
|
FCH domain only 2
|
|
chr5_-_16561936
|
0.675
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr19_+_1157462
|
0.673
|
|
STK11
|
serine/threonine kinase 11
|
|
chr14_+_101346032
|
0.669
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr14_+_59786047
|
0.669
|
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr20_-_33705244
|
0.663
|
NM_003915
|
CPNE1
|
copine I
|
|
chr20_+_43943254
|
0.663
|
NM_080603
|
ZSWIM1
|
zinc finger, SWIM-type containing 1
|
|
chr2_-_192420168
|
0.662
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr2_-_189752575
|
0.654
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr7_-_112366962
|
0.651
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr1_-_159459999
|
0.645
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr14_+_101345878
|
0.645
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr1_+_225194582
|
0.643
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr1_+_225194616
|
0.632
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr8_-_28299850
|
0.630
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr1_+_87567164
|
0.627
|
|
LMO4
|
LIM domain only 4
|
|
chr17_+_64922420
|
0.625
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr16_+_73590419
|
0.619
|
|
ZNRF1
|
zinc and ring finger 1
|
|
chr4_+_124537334
|
0.617
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_+_225194560
|
0.616
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr14_+_23654307
|
0.604
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr1_+_225194622
|
0.601
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chrX_+_146801355
|
0.599
|
|
|
|
|
chr1_+_150753601
|
0.594
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr18_+_64616296
|
0.593
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr2_+_68815416
|
0.592
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr10_-_21226536
|
0.589
|
NM_006393
|
NEBL
|
nebulette
|
|
chr3_+_115099007
|
0.585
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr2_+_151974849
|
0.585
|
NM_001177665
|
RIF1
|
RAP1 interacting factor homolog (yeast)
|
|
chr2_+_196229715
|
0.582
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr7_+_155129904
|
0.580
|
|
RBM33
|
RNA binding motif protein 33
|
|
chr2_-_128358903
|
0.577
|
NM_031445
|
AMMECR1L
|
AMME chromosomal region gene 1-like
|
|
chr6_-_42124361
|
0.576
|
|
CCND3
|
cyclin D3
|
|
chr11_-_5247948
|
0.576
|
NM_005330
|
HBE1
|
hemoglobin, epsilon 1
|
|
chr3_+_69895310
|
0.576
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_+_95595419
|
0.574
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr2_+_61258263
|
0.571
|
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast)
|
|
chr2_+_170149092
|
0.571
|
NM_004792
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr6_+_12398514
|
0.569
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr8_+_70567581
|
0.564
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr17_-_36719030
|
0.562
|
NM_001146182
|
KRTAP16-1
|
keratin associated protein 16-1
|
|
chr2_+_201689174
|
0.556
|
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr2_-_101457596
|
0.556
|
NM_001145664
|
RFX8
|
regulatory factor X, 8
|
|
chr14_-_34414603
|
0.547
|
NM_013448
NM_182648
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chr1_+_87567136
|
0.546
|
|
LMO4
|
LIM domain only 4
|
|
chr17_+_36665161
|
0.542
|
NM_030975
|
KRTAP9-9
|
keratin associated protein 9-9
|
|
chr19_-_42880184
|
0.541
|
|
ZNF607
|
zinc finger protein 607
|
|
chr12_+_100198026
|
0.540
|
NM_014503
|
UTP20
|
UTP20, small subunit (SSU) processome component, homolog (yeast)
|
|
chr2_+_198089036
|
0.539
|
|
MOBKL3
|
MOB1, Mps One Binder kinase activator-like 3 (yeast)
|
|
chr17_+_54652609
|
0.529
|
NM_001165993
NM_001165994
NM_182569
|
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1
|
|
chr2_+_120487072
|
0.525
|
NM_001184937
NM_001184939
NM_020909
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5
|
|
chr8_+_70567634
|
0.525
|
|
SULF1
|
sulfatase 1
|
|
chr10_-_90601655
|
0.524
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr2_+_120487076
|
0.523
|
NM_001184938
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5
|
|
chrX_+_146801142
|
0.523
|
NM_001185075
NM_001185076
NM_001185081
NM_001185082
NM_002024
|
FMR1
|
fragile X mental retardation 1
|
|
chr21_+_29593098
|
0.522
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr17_+_38916895
|
0.521
|
|
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8
|
|
chr5_+_137701122
|
0.516
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr8_-_13018123
|
0.516
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr17_+_38916758
|
0.515
|
NM_004941
|
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8
|
|
chr21_+_29593088
|
0.514
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chrX_+_146801236
|
0.511
|
|
FMR1
|
fragile X mental retardation 1
|
|
chr7_+_116238309
|
0.510
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr21_+_29592984
|
0.510
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr5_+_137701601
|
0.507
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr8_+_21833111
|
0.507
|
NM_015024
|
XPO7
|
exportin 7
|
|
chr20_-_1257809
|
0.507
|
NM_080489
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr6_-_50039776
|
0.507
|
NM_001037499
|
DEFB114
|
defensin, beta 114
|
|
chr1_+_87569959
|
0.505
|
|
LMO4
|
LIM domain only 4
|
|
chr11_-_46572160
|
0.500
|
|
AMBRA1
|
autophagy/beclin-1 regulator 1
|
|
chr6_-_33862690
|
0.499
|
NM_001143944
|
LEMD2
|
LEM domain containing 2
|
|
chr20_-_33783409
|
0.497
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr2_-_177210904
|
0.496
|
|
LOC375295
|
hypothetical LOC375295
|
|
chr6_-_105734535
|
0.493
|
NM_022361
|
POPDC3
|
popeye domain containing 3
|